The molecular robot
15 May 2010 - Making It Move III
 This blog has a frequent topic called Making It Move that has been featuring thus far floating camphor boats, synchronized swimming droplets and brave droplets taking on a maze. Thanks to a group of researchers from Arizona State University / University of Michigan /Caltech / Columbia University we can now add to our list molecular DNA robots moving around on a DNA platform (Lund et al. DOI). The robot in question looks like a spider with a streptavidin body and three deoxyribozyme legs derived from a DNAzyme called 8-17 DNA (used often in DNA computing).
This blog has a frequent topic called Making It Move that has been featuring thus far floating camphor boats, synchronized swimming droplets and brave droplets taking on a maze. Thanks to a group of researchers from Arizona State University / University of Michigan /Caltech / Columbia University we can now add to our list molecular DNA robots moving around on a DNA platform (Lund et al. DOI). The robot in question looks like a spider with a streptavidin body and three deoxyribozyme legs derived from a DNAzyme called 8-17 DNA (used often in DNA computing).
The robot arena is made from a single strand of DNA (7200 nucleotides) origami folded into a 2-dimensional platform measuring 65 nm x 390 nm x 32 nm with the aid of staple strands. Each spider leg on contact with a substrate site (a oligonucleotide) at the platform , catalytically cleaves it at the ribose unit. As each leg has higher substrate affinity than product affinity it will favor spots on the platform not yet trampled upon and a walking movement ensues.
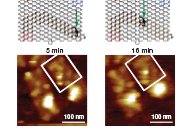
The staple strands are fitted with probes that stick out from the surface serving special purposes: they form START (complementary to the TRIGGER), TRACK (substrate) and STOP (uncleavable substrate analogs) sites. A TRIGGER oligonucleotide sets the spiders in motion with an average speed of 180 nanometer per hour. The article is accompanied by a set of very fuzzy AFM images revealing the spider movements that could just as easy be taken as evidence for the Loch Ness monster. Also included are position-time trajactories obtained from total internal reflection fluorescence microscopy which required fluorophore labeling of spiders and STOP sites.
There is always the possibility that a spider dissociates all three legs simultaneously and makes a jump to a new section of the platform. As no spiders were found at so-called CONTROL sites (a STOP site well off a track) we should be reassured this does not happen.